
Multiple sequence alignment (MSA) is a prerequisite for virtually all comparative
sequence analyses, including phylogeny reconstruction, functional motif or domain characterization,
sequence-based structural alignment, inference of positive selection, and profile based homology
searches. All such analyses take the MSA input for granted, regardless of uncertainties in the
alignment. Since errors in upstream stages tend to cascade downstream, alignment errors are an important
concern in molecular data analysis.
The GUIDANCE web-server is a powerful and user-friendly tool for assigning a confidence score for each
residue, column, and sequence in an alignment and for projecting these scores onto the MSA [1]. The server points to columns and sequences that are unreliably aligned and
enables their automatic removal from the MSA, in preparation for downstream analyses.
Three algorithms for quantifying MSA uncertainties are implemented in the server. The GUIDANCE score is
based on the robustness of the MSA to guide-tree uncertainty and relies on the bootstrap approach [2]. The Heads-or-Tails (HoT) score measures alignment uncertainty due to
co-optimal solutions [3].
GUIDANCE2 is an integrative methodology that accounts for: (1) uncertainty in the process of indel
formation;
(2) uncertainty in the assumed guide tree (as GUIDANCE); (3) co-optimal solutions in the pair-wise
alignments,
used as building blocks in progressive alignment algorithms (as HoT).
GUIDANCE is meant to be used for weighting, filtering or masking unreliably aligned
positions in sequence alignments before subsequent analysis. For example, align codon sequences
(nucleotide sequences that code for proteins) using PAGAN, remove columns with low GUIDANCE scores, and
use the remaining alignment to infer positive selection using the branch-site dN/dS test. Other analyses
where GUIDANCE filtering could be useful include phylogeny reconstruction, reconstruction of the history
of specific insertion and deletion events, inference of recombination events, etc.
GUIADNCE2 also provides a set of alternative alignments which can be used when adopting
statistical point of view, i.e. performing statistical analyses that rely on many possible alignments
that
are supported by the data.
GUIDANCE cannot tell you which alignment is better. For example, align the same
sequences using either PRANK or MAFFT and assign GUIDANCE scores to both. If the PRANK alignment has an
average score of 0.8 while the MAFFT alignment got 1 this does not mean that the MAFFT alignment is more
accurate. GUIDANCE measures the robustness of the alignment, so a perfect score means that MAFFT will
always consistently aligns the sequences in the same way, regardless of perturbations in GUIDANCE.
Still, this one way may be consistently wrong. So GUIDANCE cannot be used to choose between alternative
alignments. It can only be used to evaluate one given alignment by a certain alignment program and
identify columns where this aligner is less confident relative to other columns in the same alignment.
GUIDANCE is also not appropriate to evaluate an alignment produced by a different approach from
the ones supported in GUIDANCE (MAFFT, MUSCLE, PRANK, PAGAN and CLUSTALW). For example, you should not
run GUIDANCE on an alignment produced by T-COFFEE. Also, do not upload to GUIDANCE an alignment that you
corrected manually, even if it was originally produced by one of the supported aligners. Similarly,
alignments that used special features (e.g. MAFFT alignment that uses RNA structure information) cannot
be evaluated by GUIDANCE. In general, we recommend to always upload the sequences un-aligned and avoid
using the option to upload aligned sequences.
GUIDANCE scores reflect the robustness of an alignment to perturbations.
For this goal, a standard MSA is first generated, hereby termed "base
MSA". The user may choose between ClustalW [4],
MAFFT (the FFT-NS-1 variant) [5], and PRANK [6]. The main idea
behind the GUIDANCE2 and GUIDANCE methodologies is to construct a set of MSAs. GUIDANCE uses bootstrap
trees as guide-trees to the alignment algorithm, and compare them to the base MSA in order to estimate
its confidence level (Figure 1). Similarly, GUIDANCE2 uses bootstrp trees, vary the gap penalty score of
the alignment program scoring scheme, and employs HoT methodology (see details below).
Comparing the base alignment to the set of alternative alignments results in scores between 0-1 for each
residue, residue-pair, column and sequence of the MSA.
An in-depth description of the algorithm behind GUIDANCE can be found
in ref. [2].
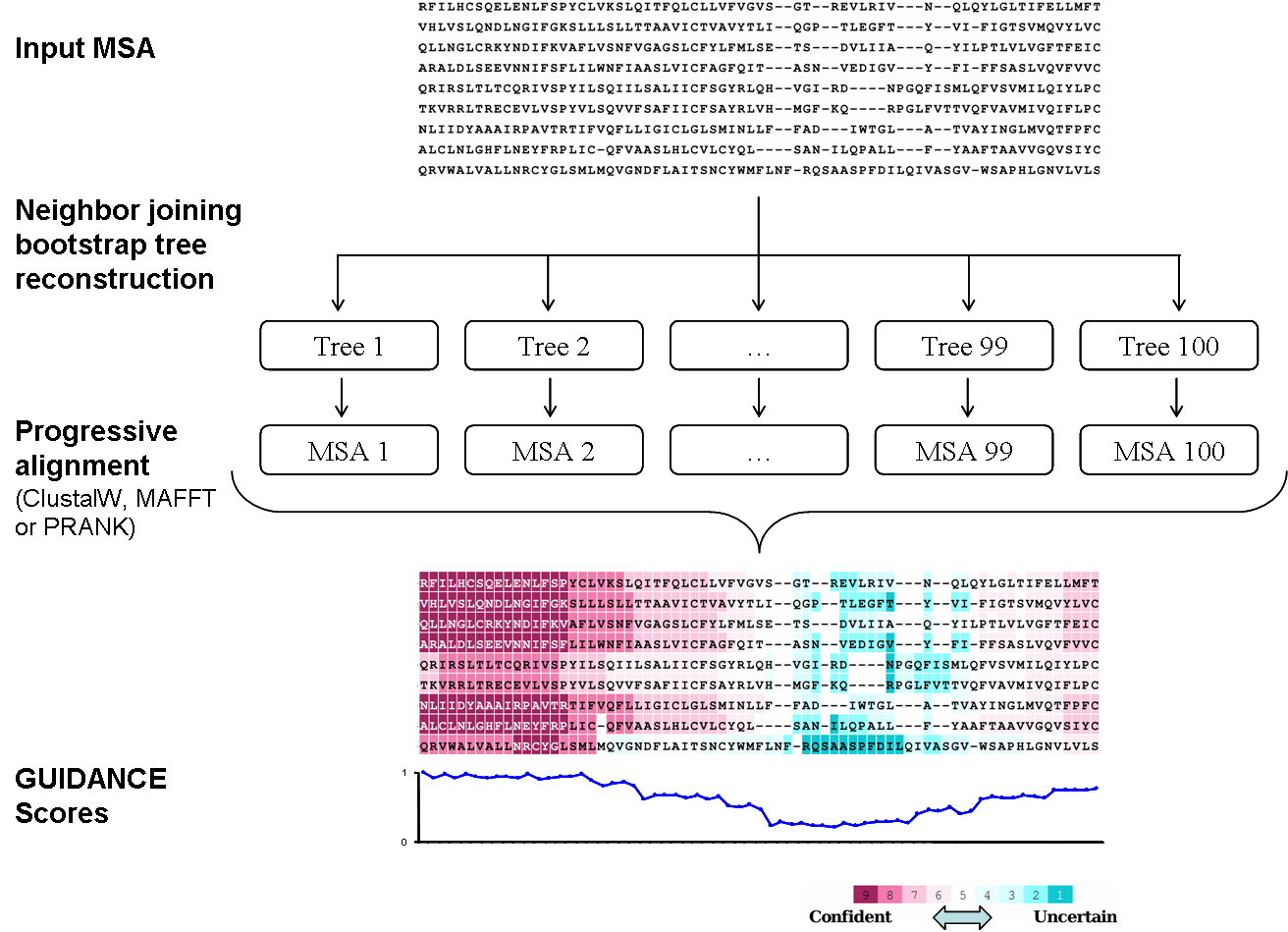
FIGURE 1
A schematic flowchart of the GUIDANCE algorithm. A base MSA is produced by any progressive alignment
method. Bootstrap neighbor joining (NJ) trees are reconstructed and given as guide trees to the
progressive alignment program, producing a set of MSAs. GUIDANCE scores are then calculated by
comparing each MSA to the base MSA, and are color coded on each residue in the alignment.
HoT (Heads-or-Tails) scores measure the alignment uncertainty by
generating a set of co-optimal MSAs and comparing them to the standard
alignment. Co-optimal MSAs are a set of alignments that are given the
same maximal score by the alignment algorithm. The co-optimal MSAs set
is constructed by reversing the sequences at each of the
pairwise-profiles-alignment steps of the progressive alignment algorithm
[3]. The comparison results in scores between 0-1 for each residue, residue-pair,
column and sequence of the MSA.
Running time depends on the dataset size (number and length of
sequences) and (for GUIDANCE2 and GUIDANCE scores) on the number of bootstrap
repeats. The major component of the running time is the multiple
alignment program used, thus MAFFT runs will be fastest and PRANK runs
slowest. In Figure 2, a comparision between the running time of GUIDANCE2 and other MSA reliability
methods when using MAFFT as the alignment method.
Note that GUIDANCE2 and GUIDANCE were run
with the default 100 bootstrap repeats, but this number can be reduced
to shorten the running time. HoT running time depends on the number of
branches in the guide tree, which increases linearly with the number of
sequences. Zorro, TCS, ALiScore, TrimAl and NOISY were run on a pre-calculated MSA.
Please note that the stand-alone versions of GUIDANCE and GUIDANCE2 support parallel computing,
thus a significant reduction in running times is possible.
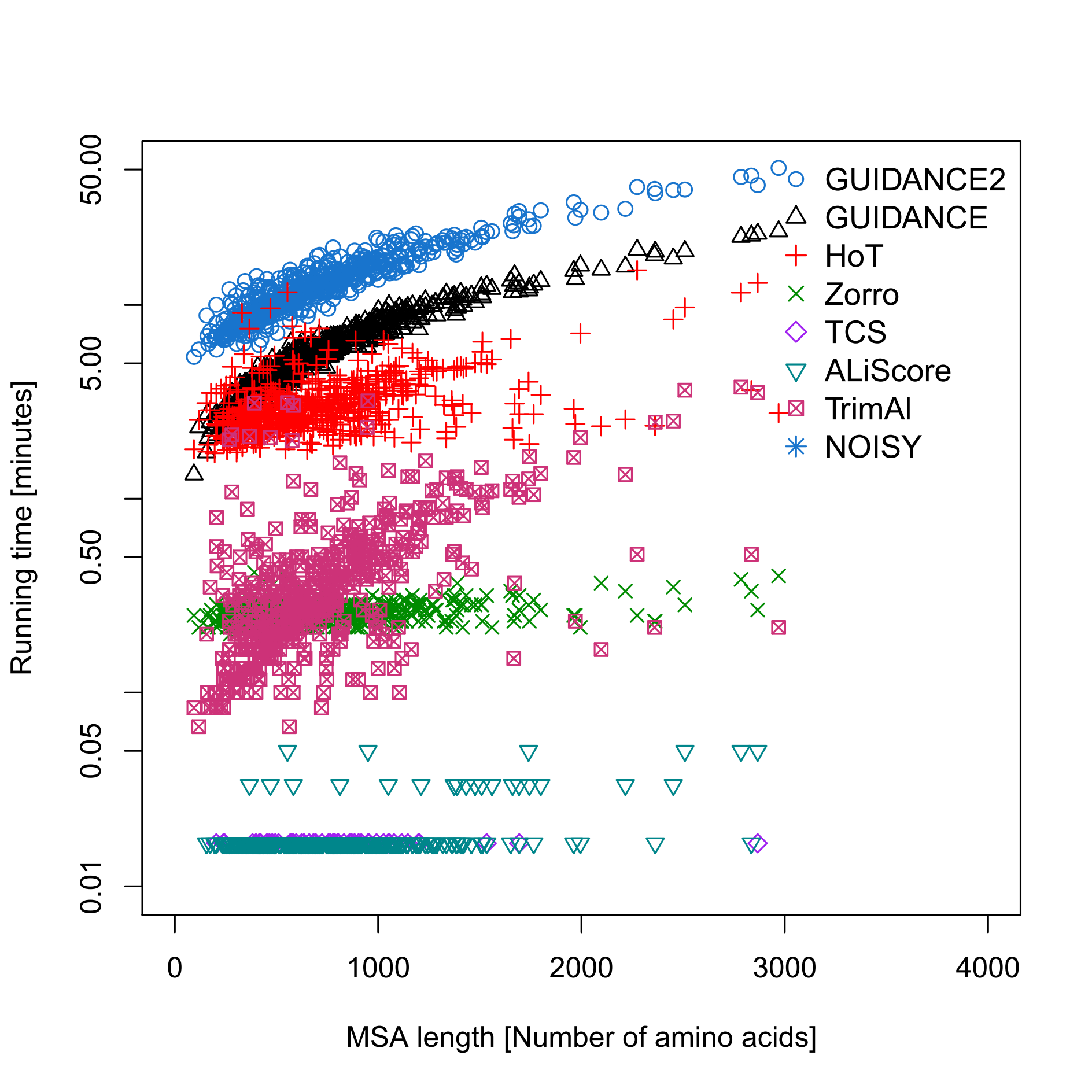
FIGURE 2: Time performance as a function of the sequence length.
Sets of 40 simulated protein sequences with different lengths were aligned
using MAFFT and analyzed by alignment reliability methods.
GUIDANCE directs you to a web page called "GUIDANCE Job Status
Page". This web page is automatically updated every 30 seconds, showing
messages regarding the different stages of the server activity. When the
calculation finishes, several links appear. For simplicity, we only
describe the output of the GUIDANCE method. Similar output is produced
by the HoT method, also implemented in this server. (for an example
output page click here)
Among other benchmarks used to evaluate the performance of GUIDANCE2 we used a benchmark of 541 simulated protein sequences. Sequences were simulated using INDELible. In order to have realistic parameters for the simulations, we first selected 541 MSAs from the OrthoMaM database, for which CDS are available for all 40 mammals included in the database. This parameter setup resulted in MSAs similar to OrthoMaM alignments (visual comparison of alignment length, number and length of indels). The control files for INDELible and the resulted simulated MSAs used as benchmark can be downloaded here.